There are no items in your cart
Add More
Add More
| Item Details | Price | ||
|---|---|---|---|
DNA sequencing is one of the most powerful tools in modern biology. It allows scientists to read the precise order of nucleotides (A, T, G, and C) in a DNA molecule, unlocking genetic information that drives life. Over the years, DNA sequencing has evolved from labor-intensive manual techniques to ultra-fast, high-throughput systems that can decode entire genomes in hours.
Let’s understand the key differences between the major DNA sequencing techniques — their principles, workflows, strengths, and limitations.
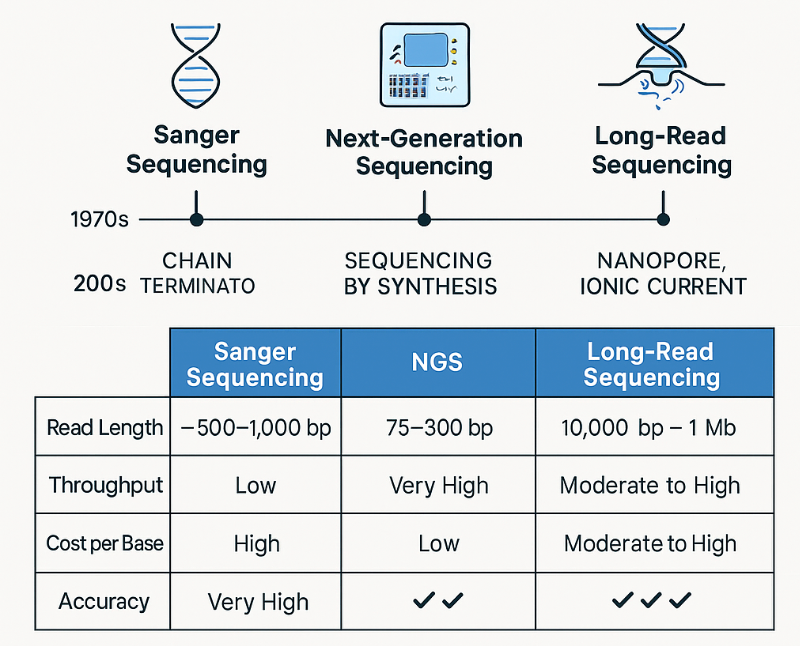
Sanger sequencing is the earliest method used to read DNA, developed by Frederick Sanger in the 1970s.
It works like reading a sentence one letter at a time, using a special trick — it adds “stop” chemicals (called dideoxynucleotides or ddNTPs) that randomly end the copying of DNA at different points.
When these fragments are separated and detected, the sequence of letters (A, T, G, C) can be read in order.
It’s slow and can only read a small section at once, but it’s extremely accurate — often used to double-check important results or confirm mutations in genes.
NGS appeared in the mid-2000s and completely changed the field.
Instead of reading one piece of DNA at a time (like Sanger), NGS can read millions of DNA fragments simultaneously.
Each fragment is copied and read base-by-base using light signals or pH changes.
Computers then reassemble all these small fragments to produce the full sequence.
It’s like having a million people reading different paragraphs of a book at once, then using a computer to join them all together.
It is massively faster and cheaper than Sanger; ideal for sequencing whole genomes, transcriptomes (RNA-Seq), or microbial communities.
Third-generation sequencing (like PacBio or Oxford Nanopore) takes another leap by reading single DNA molecules directly, without needing to make copies.
It can read very long stretches — thousands to even a million bases in one go.
These methods detect either light pulses (PacBio) or electrical signals as DNA passes through a tiny nanopore (Oxford Nanopore).
It’s like using a scanner that reads entire chapters instead of just sentences — faster and more complete.
It's best for finding large structural changes, understanding complex genomes, or assembling new genomes from scratch.
Some devices are even portable, allowing scientists to sequence DNA in the field.
| Method | Year | Principle | Strength | Limitation |
| Sanger | 1970s | Chain-termination; read one fragment at a time | Very Accurate | Slow, low throughput |
| NGS |
2000s | Millions of short reads in parallel | High throughput, low cost per base | Short reads, needs heavy computation |
| Third-Gen (Long-Read) | 2010s | Real-time single-molecule reading | Very long reads, structural insights | Higher error rate, expensive setup |
Choosing the right technique for your project depends on the a few questions that you might need to ask yourself, such as:
Understanding the differences in DNA sequencing techniques empowers you to match the right method to your research needs. While Sanger sequencing remains a gold standard for small-scale validation, NGS unlocked large-scale genomics, and long-read technologies now push boundaries for resolving complex genetic regions. Whether you're building biotech pipelines or planning experiments, picking the appropriate sequencing method is crucial to success.

Arti Rani
Educator @ Baayo